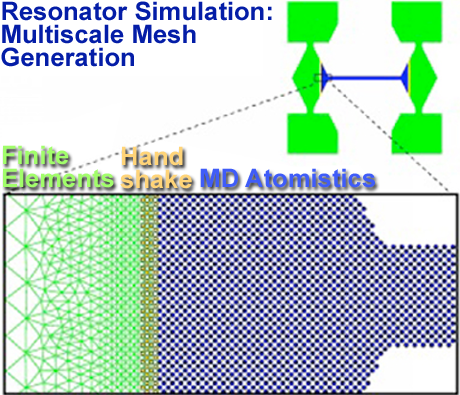
The mechanical behavior of materials often is governed by processes at multiple length scales. The challenge that this poses is particularly acute when the scales are so strongly coupled that they must be treated concurrently. Of the systems we have studied using these techniques, two are particularly interesting: the oscillations of sub-micron silicon resonators and the ductile failure of copper and other fcc metals at high strain rate. The resonators are of interest because they are used as the mechanical components in Micro-Electro-Mechanical Systems (MEMS), and departure from the conventional laws of continuum mechanics at small length scales has posed a problem for effective design. The modeling of dynamic fracture at small scales is relevant to the current generation of spallation experiments. In both cases, the overall behavior is determined by the interplay between physics at the Angstrom, nanometer and micron scales. In order to model all of the relevant scales, we have developed a technique which unifies molecular dynamics (MD) with finite elements (FE), or a nanoscale generalization we have developed known as Coarse-Grained Molecular Dynamics (CGMD). In the hybrid FE/MD approach, a handshaking region is used to couple the atomistic region to the finite element region. The coordinates in both regions are taken to be positions (displacements), so that these basic variables are well defined in the handshaking region. In the MD and FE regions, the system evolves according to the usual MD and FE equations of motion. In the handshaking region, a mean force Hamiltonian is used to transition between the two smoothly and to provide the required coupling. In CGMD, this coupling is derived explicitly from the atomistic model in order to ensure that the transition is entirely seamless. Both approaches capture the relevant physical effects at different length scales simultaneously. hton and coworkers is the scale dependence of the Young's modulus of a beam (a related recent result is shown below).
Selected Publications
- R.E. Rudd and J.Q. Broughton, "Coarse-grained molecular dynamics: Nonlinear finite elements and finite temperatures," Phys. Rev. B 72, 144104 (2005). cond-mat/0508527
- R.E. Rudd, "Coarse-grained molecular dynamics for computer modeling of nanomechanical systems," Intl. J. on Multiscale Comput. Engin. 2, 203-220 (2004)
- R.E. Rudd, "Coarse-Grained Molecular Dynamics: Dissipation due to Internal Modes," Mat. Res. Soc. Symp. Proc. 695, T10.2.1-6 (2002).
- R.E. Rudd, "Concurrent Multiscale Modeling of Embedded Nanomechanics," Mat. Res. Soc. Symp. Proc. 677, AA1.6.1-12 (2001).
- R.E. Rudd and J.Q. Broughton, "Concurrent Coupling of Length Scales in Solid State Systems," Phys. Stat. Sol. (b) 217, 251 (2000).
- R.E. Rudd and J.Q. Broughton, "Atomistic Simulation of MEMS Resonators through the Coupling of Length Scale," J. Modeling and Simulation of Microsystems 1, 29 (1999).
- R.E. Rudd and J.Q. Broughton, "Coarse-grained molecular dynamics and the atomic limit of finite elements," Phys. Rev. B 58, R5893 (1998).