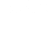The Cooperative Research Center for NanoScaffold-based Chlamydia trachomatis Vaccines
- Home
- Research and Development
- Biosciences & Biotechnology
- Center for Chlamydia Trachomatis Vaccines
Developing new chlamydial vaccines supported by nanotechnology
Our multidisciplinary team of leading experts in immunology and nanotechnology are developing and testing a new type of vaccine to prevent sexually transmitted infections caused by the Chlamydia trachomatis (Ct) pathogen, providing a viable pathway to address this widespread public health threat.
Overview
Ct affects millions of people every year
Ct, the bacterium that causes chlamydia, is the most common sexually transmitted pathogen in the world, affecting more than 130 million people every year. When left untreated, Ct infection can lead to pelvic inflammatory disease, chronic pelvic pain, ectopic pregnancy, and infertility. It is also the most common cause of preventable blindness worldwide, and it has been linked to ovarian cancer.
Despite considerable efforts to develop a Ct vaccine, to date, there are no vaccines that can protect against this widespread public health threat. Previous studies have shown that immunization with a Ct membrane protein can provide significant protection against infection if its native structure is preserved. However, formulating and delivering this type of vaccine remains a major hurdle.
Nanolipoproteins offer unique vaccine development and delivery options
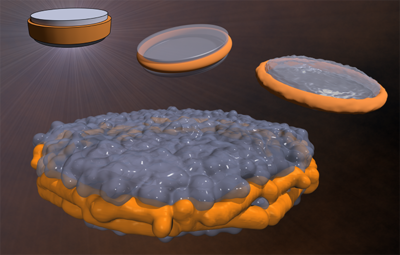
A novel technology—called nanolipoprotein (NLP) particles—provides a unique approach to vaccine development and delivery. NLPs are small structures produced by mixing scaffold proteins and lipids, which self-assemble into a disc-shaped (nanodisc) structure and provide a stable platform where other biomolecules, such as vaccine elements, can be attached.
NLPs are highly customizable, making them an ideal platform for vaccine formulation, as well as an effective tool for in vivo vaccine delivery via various routes. Their tiny size makes it possible for them to take advantage of natural pathways into cells, particularly immune cells relevant to vaccine delivery.
LLNL researchers have been developing and refining the platform for more than a decade, focusing on its ability to serve as an immunogenetically inert scaffold for drug and vaccine delivery. Attaching vaccine elements, including antigens and adjuvants, to NLPs results in vaccines that elicit a stronger, more targeted immune response than when the components are administered separately.
A Center to leverage NLPs for Ct vaccines
In 2019, the National Institutes of Health (NIH) established, through its National Institute of Allergy and Infectious Diseases, the Cooperative Research Center for NanoScaffold-based Chlamydia trachomatis Vaccines, providing an initial five years of funding.
Center researchers will leverage NLPs to explore solutions to both the development and the delivery of an effective vaccine.
First, they will identify the most promising antigen formula to protect against the Ct pathogen and explore how to administer it via an NLP delivery platform that preserves the molecule’s structure as it is delivered.
Then, they will test the vaccine, establishing the foundation needed to move the work toward clinical trials.
The Center takes advantage of expertise from three institutions:
- University of California (UC), Irvine researchers, including a leading expert in chlamydia biology, will focus on developing models that mimic the response of chlamydia human infections.
- Using these models, LLNL scientists, who developed the nanotechnology platform, will refine NLP formulations for use in Ct vaccine development and delivery.
- UC Davis health researchers will devote their efforts to the safety and efficacy of the vaccine formulations.
The collaboration could result in a nanotechnology delivery platform that will form the basis for future efforts to deliver drugs and vaccines to fight other infectious diseases.
Projects
Our work is organized into three research projects that tackle diverse approaches to vaccine development.
Project 1: Develop candidate antigens using membrane proteins
Our Project 1 focus is vaccine development. Our approach includes engineering a robust Ct vaccine using two protective antigens found in surface-exposed proteins within the Ct outer membrane—proteins that connect the outside world to the intracellular world.
One antigen candidate is the major outer membrane protein (MOMP). Studies have shown that MOMP-based immunizations can induce protection against infection and disease in animal models if its native structure is preserved. Our approach uses cell-free synthesis to generate a correctly-folded protein structure.
Another candidate is a group of surface-exposed candidate antigens known as polymorphic membrane proteins (PMPs). Initial studies indicate that PMPs may help broaden the protective immune responses elicited by MOMP. However, successful use of full-length PMPs for vaccine applications require a delivery platform that can support its use.
With this in mind, our research team will use synthetic biology to select and produce protein sequences that we can test to determine the sequences that will make the most effective vaccines. We will embed the membrane proteins onto nanoscaffolds to provide a stable platform.
Team
- Matt Coleman, Principal Investigator, LLNL
- Sean Gilmore, LLNL
- Mimi Yung, LLNL
Project 2: LLNL LDRD research project: Delivering RNA vaccines using nanoparticles
The focus of our Laboratory Directed Research and Development (LDRD) project is to enhance the efficacy of delivering nucleic acid vaccines. Our approach leverages the capability of messenger RNA (mRNA)-based vaccines that can self-replicate in host cells, reducing the dose needed to achieve sufficient antigen expression and the desired immune response. As a proof of concept, we will develop approaches to deliver mRNA encoding to the Ct antigens in vivo.
The self-amplifying mRNAs, known as SAMs or replicons, harness the host cell’s ability to produce the encoded antigen. Our team has already investigated ways to tailor our NLP particles to bind and package large mRNA molecules that encode key vaccine antigens, thereby facilitating their intracellular delivery and protein expression. Building on this capability, we will develop and evaluate various NLP compositions coupled with replicons encoded with the Ct antigens to identify the most promising formulations.
In addition to achieving the desired immune response by harnessing the host cell’s ability to produce antigens, our approach also avoids costly and time-consuming protein-based antigen development. Instead, as the encoded mRNA actively propagates, the antigen is expressed at a high level.
Team
- Nicholas Fischer, Principal Investigator, LLNL
- Wei He, LLNL
- Sandra Peters, LLNL
- Amy Rasley, LLNL
- Dina Weilhammer, LLNL
Project 3: Validate the safety and efficacy of vaccine candidates in vivo
Our Project 3 focus is validating the safety and efficacy of our candidate Ct vaccines in vivo. We anticipate that through these activities, we will be able to refine, and possibly combine, formulations developed in Project 1 and Project 2.
Team
- Luis de la Maza, Principal Investigator, UC Irvine
- Sukumar Pal, UC Irvine
Supporting cores
Three supporting cores will enable Center investigators to use state-of-the-art methodologies, including protein production, in their research projects as well as bioinformatic and immunological analyses.
Core 1 provides an integrated set of tools to support biostatistical and bioinformatics design, analysis, and data sharing across the research projects. Bioinformatic analysis will support Project 1 and Project 2, while all three research projects will use statistical analyses for hypothesis support and significance testing.
Team
- Patrik D’haeseleer, Principal Investigator, LLNL
- Laurel Beckett, UC Davis
Core 2 provides facilities, instrumentation, and storage necessary for production, characterization, preservation, and distribution of NLP particles and protein antigens. The Core 2 team produces plasmids needed for cell-free translation within Project 1, as well as purified proteins and complexed NLPs for vaccine testing.
Team
- Brent Segelke, Principal Investigator, LLNL
- Joseph McKeown, LLNL
- Megan Shelby, LLNL

Core 3 provides immunological analyses and safety/pathology services. The Core 3 team works closely with Project 1 and Project 2 to provide quantitative data regarding the immune response initiated by vaccine candidates. In addition, the Core 3 team characterizes the immune responses induced in mice, supporting the work done by the Project 3 research team.
Team
- Amy Rasley, Principal Investigator, LLNL
- Sandy Borowsky, UC Davis
- Dina Weilhammer, LLNL
Timeline
Our initial research plan focuses on developing and testing a safe and effective Ct vaccine that overcomes the limitations of current vaccine efforts and leverages LLNL-developed nanotechnology.
- Years 1–3: Formulation development and application
- Years 2–4: Safety
- Years 3–5: Efficacy
By the end of the Center’s first five years, we anticipate that our research team will establish the foundation needed to move the work toward industrial testing and clinical trials and be poised to provide a critical technology path forward for addressing other vaccine needs.
News and Publications
- Information for the 28th Annual Principles of STI/HIV Research and Public Health Practice Course Now Available (University of Washington, February 2020)
- NIH looks to Lab to help develop chlamydia vaccine (LLNL news, October 2019)
- A Long Shot Pays Off in Vaccine Development (Science & Technology Review, March 2019)
Our Team
The Center is a collaboration between LLNL scientists, who developed the nanotechnology platform, and researchers at two University of California (UC) campuses, including a leading expert in chlamydia biology from UC Irvine and experts from UC Davis who will focus on studying the safety and efficacy of the vaccine formulations.
Center directors
Other team members
External Review Committee
The Center’s External Review Committee provides advice related to the progress of the Center’s research aims, including opportunities for modification or expansion of the research plan.
Karen Elkins
U.S. Food and Drug Administration
Jeff Fairman
SutroVax
Michael Jewitt
Northwestern University
Raymond Johnson
Yale School of Medicine
Dorothy Patton
University of Washington
Work With Us
Developmental Research Program
The Developmental Research Program (DRP) is aimed at early career investigators and other scientists who are interested in receiving funding for pilot projects that last up to two years, on topics related to the Center’s research objectives.
Current DRP projects include:
- A native outer membrane vesicle vaccine for prevention of gonococcal diseases, Peter Beernink, University of California, San Francisco School of Medicine
- Extraction and purification of the Chlamydia trachomatis serovars D and E native major outer membrane protein, Anatoly Slepenkin, University of California, Irvine
We anticipate issuing annual requests for applications during each year that the Center is funded. DRP projects will be funded for 1–2 years, with maximum funding of $30K per year. The second year of funding will be based on satisfactory progress on research goals and milestones. It is our hope that the funded projects will be able to obtain preliminary data that will support follow-up funding in collaboration with the Center.
Requests for DRP funding will be evaluated by scientists, clinicians, and subject matter experts using a two-tier review process:
- Tier 1: A scientific peer review of proposals against established criteria for determining scientific merit by selected outside reviewers not associated with the Center.
- Tier 2: A programmatic review within the Center that compares submissions and recommends proposals for funding based on scientific merit and relevance to the overall goals of the Center’s research program.
Applications will be evaluated based on the significance/rationale for the proposed study, its innovation/novelty, the feasibility of the experimental plan, and the potential for translation to Center deliverables. Special consideration will also be given to studies that fund new investigators, with no more than six years of experience as principal investigator or senior scientist.
Please return to this page for future information about DRP applications.
Career Opportunities
Faculty Mini Sabbatical Program at LLNL
Science Undergraduate Laboratory Internships